Golden Gate assembly of the β-carotene pathway
Teams were tasked to build a β-carotene pathway in yeast. To do this, they were given a range of 4 genes encoding a β-carotene pathway. Each gene had a choice of 6 promoters with a range of outputs (1 = no promoter/CDS, promoter 2 = strongest, promoter 7 = weakest). Teams decided on how to build the pathway, using defined or combinatorial promoters for each gene. Using an Echo to put together each team’s reaction, pathways were assembled into a 2μ plasmid using Golden Gate assembly in the YTK format.
Here are some of the results:
Group 1, Jane and Fabien

Group 4, David and Emilio

Group 6, Dimitra and Maarten
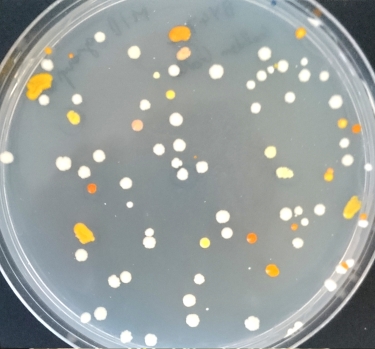
Group 8, Zuzana and Amritpal

Group 12, Sandra and Elizabeth
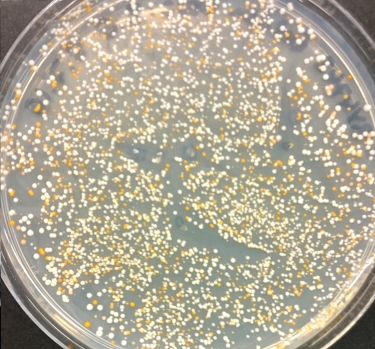
Group 15, Matt and Steph

SCRaMbLEing pigment production
One of the major design features of the Sc2.0 project is the insertion of loxPsym sites in the 5’ UTR of non-essential genes. This allows for massive Cre recombinase-mediated rearrangement of the synthetic genome. This process is called SCRaMbLE, to find out more, see here.
Teams were given multiple strains to perform SCRaMbLE experiments. One of the experiments involved strains containing plasmids with 2 different β-carotene expression pathways, pathway 1 was unbalanced and overproduced a yellow intermediate (neurosporene), whilst pathway 2 produced the orange final product (β-carotene). The genes were all formatted to be interspersed with loxPsym sites so as to be potentially rearranged during SCRaMbLE. Each plasmid was present in two different strain backgrounds. One strain had a wild-type genetic background whilst the other had a synthetic chromosome V.
Each combination of pathway and strain was SCRaMbLEd by the teams in order to assess the effect of genetic background on the SCRaMBLEing of a heterologous metabolic pathway.

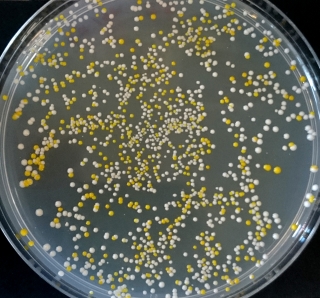

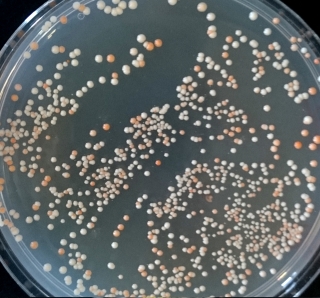
The teams found that SCRaMbLEing in a wild-type background increased the frequency of white colonies, indicating a loss of pathway function. The colonies that still had a functional β-carotene pathway in the wild-type background showed a higher variability in expression phenotype than observed with the synthetic strain background.
SCRaMbLEing in the synthetic background appeared to result in the best β-carotene producers. Some of the pathway 1 colonies appeared orange, from which we deduce that they are successfully producing β-carotene, and some of the pathway 2 colonies appeared very brightly orange. We concluded that SCRaMbLEing in a wild-type background resulted in the most variation in pathway output whilst SCRaMbLEing in a synthetic background generated strains with the best β-carotene production.
All of the plates that were left have been photographed and can be viewed here.
The handbook used on the course can be downloaded here.
